Identification of a fragment of the internal transcribed spacer region of rDNA in Cenchrus purpureus (Poaceae)
Main Article Content
Abstract
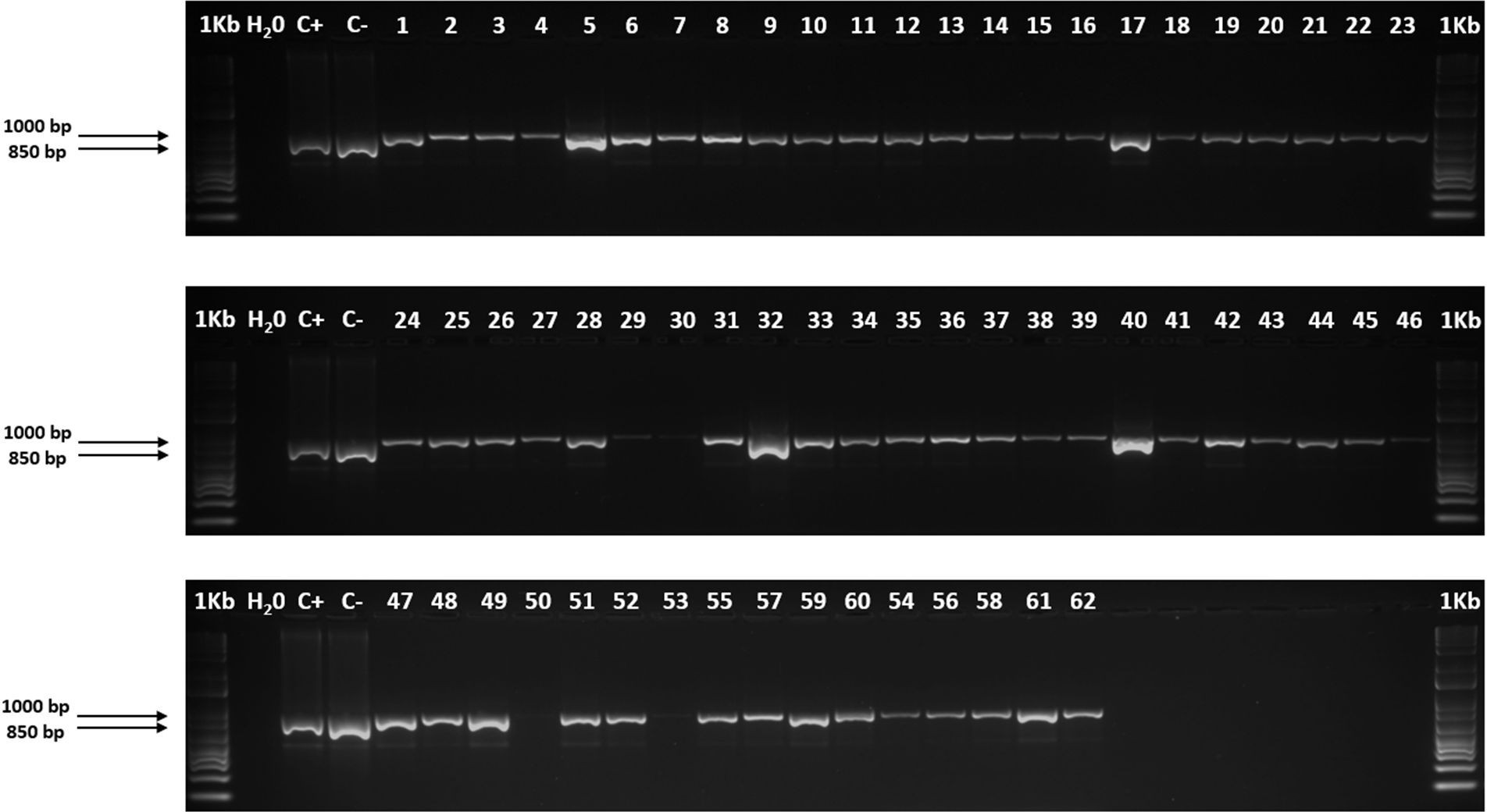
The internal transcribed spacer (ITS) region is easy to amplify due to its numerous copies present in the plant genome, but there in not researchers of this region in the forage species Cenchrus purpureus. This study aimed to identify by Polymerase Chain Reaction (PCR) a fragment of the ITS region of the ribosomal DNA of the nucleus in C. purpureus using the primers ITS1/ITS4 and compare it with two samples of Urochloa spp. The molecular study used DNA samples from 62 C. purpureus accessions from the grass and forage germplasm bank belonging to Instituto de Ciencia Animal and two samples of Urochloa spp. accessions from the Genetic Resources Program of the Alliance of International Bioversity and the International Center for Tropical Agriculture. The DNA from the 62 C. purpureus accessions and Urochloa spp. controls were amplified with ITS1/ITS4 primers. The amplification products revealed a clear polymorphic band in the gels for each accession and an approximate size of 1000 bp in C. purpureus and 850 bp in Urochloa spp. The primers determined differences in amplifications between samples of C. purpureus and Urochloa spp. although they did not show variability between accessions of the same species. These molecular markers can be used to verify PCR amplification of C. purpureus DNA samples and to differentiate species in Poaceae genera.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Those authors that have publications with this journal accept the following terms:
1. They will retain their copyright and guarantee the journal the right of first publication of their work, which will be simultaneously subject to the License Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) that allows third parties to share the work whenever its author is indicated and its first publication this journal. Under this license the author will be free of:
- Share — copy and redistribute the material in any medium or format
- Adapt — remix, transform, and build upon the material
- The licensor cannot revoke these freedoms as long as you follow the license terms.
Under the following terms:
- Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- NonCommercial — You may not use the material for commercial purposes.
- No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
2. The authors may adopt other non-exclusive license agreements to distribute the published version of the work (e.g., deposit it in an institutional telematics file or publish it in a monographic volume) whenever the initial publication is indicated in this journal.
3. The authors are allowed and recommended disseminating their work through the Internet (e.g. in institutional telematics archives or on their website) before and during the submission process, which can produce interesting exchanges and increase the citations of the published work. (See the Effect of open access).
References
Ahmadi, H., Solouki, M., Fazeli-Nasab, B., Heidari, F. & Sayyed, R.Z. 2022. Internal transcribed spacer (ITS) regions: A powerful tool for analysis of the diversity of wheat genotypes. Indian Journal of Experimental Biology, 60(2): 137-143, ISSN: 0975-1009. http://op.nis- cair.res.in/index.php/IJEB/article/view/34886.
Al Rahbawi, S.M., Al-Edhari, A.H. & Sardar, A.S. 2021. Phylogenetic Study of the Genus Phalaris L. (Poaceae) based on Nuclear Internal Transcript Region (ITS) in Iraq. Annals of the Romanian Society for Cell Biology, 25(3): 8278-8281, ISSN: 2067-8282. http://annalsofrscb.ro/in- dex.php/journal/article/view/2368.
Alaklabi, A. 2021. Ficus Species Genetic Diversity Based on Internal Transcribed Spacer (ITS) Region Analysis. Egyptian Academic Journal of Biological Sciences, H. Botany, 12(1): 21-27, ISSN: 2090-3820. https://doi.org/10.21608/eajbsh.2021.148134.
Almutairi, Z.M. 2021. Molecular identification and phylogenetics of local pearl millet cultivars using internal-transcribed spacers of nuclear ribosomal DNA. Plant Genetic Resources: Characterization and Utilization, 19(4): 339-346, ISSN: 1479-2621. https://doi.org/10.1017/S1479262121000393.
Álvarez Báez, Y. 2021. Comportamiento productivo de nuevas variedades de Cenchrus purpureus en el Valle del Cauto, Cuba. Tesis de Doctorado. Universidad de Granma. Cuba. p. 99.
Álvarez, Y., Herrera, R.S., Ramírez, J.L., Verdecia, D.M., Benítez, D. & López, S. 2024. Performance of Cenchrus purpureus varieties tolerant to salinity under the edaphoclimatic conditions of Granma province, Cuba. Cuban Journal of Agricultural Science, 58: e01, ISSN: 2079-3480. https://cu-id.com/1996/v58e01.
Baldwin, B.G., Sanderson, M.J., Porter, J.M., Wojciechowski, M.F., Campbell, C.S. & Donoghue, M.J. 1995. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden, 82(2): 247-277, ISSN: 2162-4372.
https://www.jstor.org/stable/2399880.
Bult, C. & Zimmer, E. 1993. Nuclear ribosomal RNA seguences for inferring tribal relationships within Onagraceae. Systematic Botany, 18(1): 48-63, ISSN:
-2324. https://www.jstor.org/stable/2419787.
Chen, Z.T., Huang, Q.L., Pan, W.B. & Huang, Y.B. 2010. Sequence analysis of the rDNA ITS region of Pennisetum species (Poaceae). Acta Prataculturae Sinica, 19(4): 135, ISSN: 1004-5759. http://cyxb.magtech.com.cn/EN/Y2010/V19/I4/135.
Cruz, R., Sosa, A., Herrera, R.S. & Martínez, R.O. 1993. Identificación electroforética de Pennisetum purpureum cv. King grass. Revista Cubana de Ciencia Agrícola, 27(2): 219-223, ISSN: 2079-3480. https://cjascience.com/in-dex.php/CJAS.
de Vere. N., Rich, T.C.G., Ford, C.R., Trinder, S.A., Long, C., Moore, C.W., Satterthwaite, D., Davies, H., Allainguillaume, J., Ronca, S., Tatarinova, T. Garbett, H., Walker, K. & Wilkinson, M.J. 2012. DNA Barcoding the Native Flowering Plants and Conifers of Wales. PLoS ONE, 7(6): e37945, ISSN: 1932-6203. https://doi.org/10.1371/journal.pone.0037945.
Fortes, D., Herrera, R.S. & Herrera, M. 2023. Morphoagronomic performance of Cenchrus purpureus new clones. Cuban Journal of Agricultural Science, 57: 8, ISSN: 2079-3480. https://cjascience.com/in-dex.php/CJAS/article/view/1105.
Ghosh, J. S., Bhattacharya, S. & Pal, A. 2017. Molecular phylogeny of 21 tropical bamboo species reconstructed by integrating non-coding internal transcribed spacer (ITS1 and 2) sequences and their consensus secondary structure. Genetica, 145(3): 319-333, ISSN: 1573-6857. https://doi.org/10.1007/s10709-017-9967-9.
González, A.C. 2002. Detección del polimorfismo genético mediante marcadores bioquímicos en plantas. En: Marcadores moleculares: nuevos horizontes en la genética y la selección de las plantas. Cornide, M. T. (Coord.). Ed. Félix Varela, La Habana, Cuba. pp. 36-63. ISBN: 959-258-351-X.
González, A.T. & Morton, C.M. 2005. Molecular and morphological phylogenetic analysis of Brachiaria and Urochloa (Poaceae). Molecular Phylogenetics and Evolution, 37(1): 36-44, ISSN: 1095-9513. https://doi:10.1016/j.ympev.2005.06.003.
González, C. & Martínez, R.O. 2019. Genetic characterization of clones and varieties of Cenchrus purpureus with microsatellite markers. Cuban Journal of Agricultural Science, 53(3): 307-318, ISSN: 2079-3480. https://cjas-cience.com/index.php/CJAS/article/view/914.
Herrera, R., García, M. & Cruz, A.M. 2019. Study of morphoagronomic indicators of Cenchrus purpureus clones for biomass production. Cuban Journal of Agricultural Science, 53(2): 189-196, ISSN: 2079-3480. https://cjascience.com/index.php/CJAS/article/view/886.
Herrera, R.S. 2022. Evaluation of Cenchrus purpureus varieties tolerant to drought in the western region of Cuba. Cuban Journal of Agricultural Science, 56(2): 135-143, ISSN: 2079-3480. https://cjascience.com/in-dex.php/CJAS/article/view/1049.
Hsiao, C., Chatterton, N. J., Asay, K. H. & Jensen, K. B. 1995. Molecular phylogeny of the Pooideae (Poaceae) based on nuclear rDNA (ITS) sequences. Theoretical and Applied Genetics, 90: 389-398, ISSN: 1432-2242. https://doi.org/10.1007/BF00221981.
Hsiao, C., Jacobs, S.W.L., Chatterton, N.J. & Asay, K.H. 1998. A molecular phylogeny of the grass family (Poaceae) based on the sequences of nuclear ribosomal DNA (ITS). Australian Systematic Botany, 11(6): 667-688, ISSN:
-5701. https://doi.org/10.1071/SB97012.
Jones, L., Twyford, A.D., Ford, C.R., Rich, T.C., Davies,
H., Forrest, L.L., Hart, M.L., McHaffie, H., Brown, M.R., Hollingsworth, P.M. & De Vere, N. 2021. Barcode UK: A complete DNA barcoding resource for the flowering plants and conifers of the United Kingdom. Molecular Ecology Resources, 21(6): 2050-2062. ISSN: 1755-0998.
https://doi.org/10.1111/1755-0998.13388.
Liao, H.C., Ming-Hui, C. & Chih-Hui, C. 2020. Barcode of nuclear ribosomal internal transcribed spacer regions (ITS) as a useful tool to recognize a newly naturalized and potentially invasive weed, Chloris pilosa Schumach. (Poaceae), in Taiwan. Taiwania, 65(2): 129, ISSN: 0372-333X. https://doi.org/10.6165/tai.2020.65.129.
Liston, A., Robinson, W.A., Oliphant, J.M. & Álvarez-Buylla,
E.R. 1996. Length variation in the nuclear ribosomal DNA internal transcribed spacers of non-flowering seed plants. Systematic Botany, 21(2): 109-120, ISSN: 1548-2324. https://doi.org/10.2307/2419742.
Nadeem, M.A., Nawaz, M.A., Shahid, M.Q., Doğan, Y., Comertpay, G., Yıldız, M., Hatipoğlu, R., Ahmad, F., Alsaleh, A., Labhane, N., Özkan, H., Chung, G. & Baloch, F.S. 2018. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnology & Biotechnological Equipment, 32(2): 261-285, ISSN: 1310-2818. https://doi.org/10.1080/13102818.2017.1400401.
Nguyen, B.T., Le, L.B., Pham, L.P., Nguyen, H.T., Tran,
T.D. & Van Thai, N. 2021. The effects of biochar on the biomass yield of elephant grass (Pennisetum purpureum Schumach) and properties of acidic soils. Industrial Crops and Products, 161: 113224, ISSN: 1872-633X. https://doi.org/10.1016/j.indcrop.2020.113224.
Özbek, Ö. 2024. Molecular Markers Used to Reveal Genetic Diversity and Phylogenetic Relationships in Crop Plants. OBM Genetics, 8(4): 1-25, ISSN: 2577-5790. https://doi.org/10.21926/obm.genet.2404274.
POWO. 2025. Plants of the World Online. Kew Royal Botanic Gardens. Available at: https://powo.sci-ence.kew.org/taxon/urn:lsid:ipni.org:names:77106033-1. [Consulted: January 03, 2025].
Risterucci, A., Grivet, L., N’Goran, Pieretti, J., Flament, M. & Lanaud, C. 2000. A high-density linkage map of Theobroma cacao L. Theoretical and Applied Genetics, 101: 948-955, ISSN: 0040-5752. https://doi.org/10.1007/s001220051566.
Sun, Y., Skinner, D.Z., Liang, G.H. & Hulbert, S.H. 1994. Phylogenetic analysis of Sorghum and related taxa using internal transcribed spacers of nuclear ribosomal DNA. Theoretical and Applied Genetics, 89: 26-32, ISSN: 1432-2242. https://doi.org/10.1007/BF00226978.
Tan, F., He, L., Zhu, Q., Wang, Y., Chen, C. & He, M. 2022. Pennisetum hydridum: a potential energy crop with multiple functions and the current status in China. BioEnergy Research, 15: 850-862, ISSN: 1939-1242. https://doi.org/10.1007/s12155-021-10263-7.
Wang, J., Yan, Z., Zhong, P., Shen, Z., Yang, G. & Ma, L. 2022. Screening of universal DNA barcodes for identifying grass species of Gramineae. Frontiers in Plant Science, 13: 998863, ISSN: 1664-462X. https://doi.org/10.3389/fpls.2022.998863.
Wessapak, P., Ngernsaengsaruay, C. & Duangjai, S. 2023. A taxonomic revision of Cenchrus L. (Poaceae) in Thailand, with lectotypification of Pennisetum macrostachyum Benth. PhytoKeys, 234: 1-33, ISSN: 1314-2003. https://doi.org/10.3897/phytokeys.234.106486.
White, T. J., Bruns, T., Lee, S.J.W.T. & Taylor, J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide tomethods and applications. M. Innis, D. Gelfand, J. Sninsky & T. White (eds.). Academic Press, INC. San Diego California. pp. 315-322. ISBN: 0-12-372181-4.